A new study discovers how intestinal bacteria and blood metabolites indicate an early risk of diabetes and how to diet and tailor -made exercise reverse the trend.
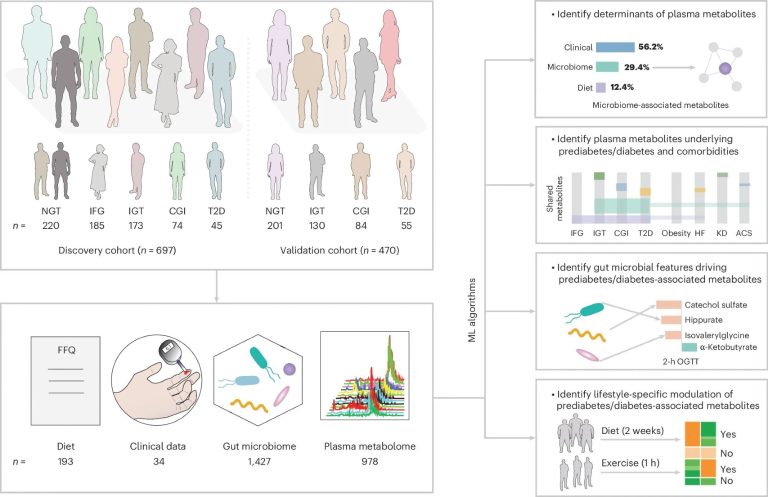
 The FBG and the OGTT were used to filter individuals with a variable degree of glucose intolerance. GBDT algorithm has been used to predict plasma metabolites based on data collected from FFQ, clinical tests and intestine microbiome profiling. N indicates the size of the sample for both cohorts, or the number of characteristics in the sets of dietary metabolome, clinical, intestinal and plasma microbiome.
The FBG and the OGTT were used to filter individuals with a variable degree of glucose intolerance. GBDT algorithm has been used to predict plasma metabolites based on data collected from FFQ, clinical tests and intestine microbiome profiling. N indicates the size of the sample for both cohorts, or the number of characteristics in the sets of dietary metabolome, clinical, intestinal and plasma microbiome.
In a recent study by the journal Nature MedicineThe researchers conducted a metabolome profiling study to study the role of microbial metabolites in prediabetes and type 2 diabetes (T2D). They used two Swedish cohorts comprising 1,167 participants aged 50 to 64 for their analyzes.
The results of the study revealed the presence of 502 blood metabolites linked to the homeostasis of altered glucose, of which 143 were associated with the human intestinal microbiome. The study highlights the role of the dynamics of microbiome-metabolome in prediabetes and T2D pathophysiology and the role of short-term lifestyle changes (diet and exercise) in the modulation of these dynamics.
Background
Type 2 diabetes (T2D) is a global public health problem, estimated at more than 830 million adults. The condition is chronic, characterized by the incapacity of the organism to adequately regulate glucose metabolism, resulting in excessively high blood sugar, which leads to complications, in particular cardiovascular diseases (CVD), renal diseases, nerve lesions and the risk of increased mortality.
Alarming, the prevalence of T2D increases at unprecedented rates, going from 200 million in 1990 to more than 830 million in 2022. Research revealed that the pathophysiology of the condition is very complicated, resulting from the interaction between genetic and environmental variables. Recent studies have suggested the in-depth role of the food and the intestinal microbiome in T2D pathogenesis, with around 70% of the T2D incidence now attributed to suboptimal regimes and their undesirable effects on intestinal bacteria.
Unfortunately, the mechanistic influence of intestinal microbial metabolites on pathogenesis and the progression of T2D remains poorly understood.
About the study
This study aims to fill these gaps in the literature by identifying intestinal microbial metabolites modulating the control of host (human) glucose and, in turn, contributing to prediabetes and T2D. The study data were obtained from two Swedish prediabetic cohorts – the altered glucose tolerance cohort (IGT; n = 697), which served as a “discovery” cohort, and the Swedish cardiopulmonary cohort (scapis; n = 470), which served as a cohort of “validation”.
The collection of the study data included glucose measures with an empty stomach in the morning, an oral glucose tolerance test of 75 g (OGTT) and a collection of samples of venous blood on an empty stomach. The results of these tests, in tandem with the criteria of the World Health Organization of 1999, were used to divide the participants in the study in five sub-groups: 1. Normal tolerance to glucose (NGT), 2.. (CGI) and 5. T2D.
The blood samples taken were subjected to a plasma metabolomic using the Metabolon platform. Microbial associated metabolites have been identified using gradient stimulation decision -making trees and random -machine (ML) random learning models.
In addition, participants had to fill the Findisc questionnaires (which reflect insulin resistance more strongly than blood sugar) and provide faecal samples, the latter were subjected to a fecal microbial profiling via metagenomics Tests.
Study results
Study participants were observed between 50 and 64 years old, with a classification of subgroups based on OGTT revealing 220 participants with NGT, 185 with IFG, 173 with IGT, 74 with CGI and 45 with T2D detected by screen. The metabolomics of the blood plasma of the participants revealed 978 plasma metabolites obtained mainly from the metabolism of lipids (45.4%) and amino acids (22.1%).
The models of decision trees (GBDT) boosted by the gradient revealed that 645 metabolites in the discovery cohort significantly associated with IFG, IGT, CGI or T2D. Among these, 502 metabolites overlooked in the meaning in the validation cohort, suggesting their role as potential biomarkers of glucose control (prediabetes and T2D biomarkers). In particular, 143 of these metabolites were associated with microbiome data and 272 with diet data.
“These results show that potential determinants persist in prediabetes and T2D, the intestinal microbiome alone representing almost a third of the variance of blood metabolite, twice measured in healthy individuals.”
It has been found that the identified metabolomic profiles overlap with previously identified signatures of prediabetes, T2D, acute coronary syndrome (ACS), heart failure (HF) and kidney disease (KD). This confirms that the dynamics of the microbiome-metabolome is disturbed before the start of the MCV, thus suggesting potential early intervention targets against the incidence of cardiometabolic diseases. For example, interactions mediated by metabolites have mediated between specific intestinal bacteria (Hominifimenecus microfluidus And Blautia Wexlerae), with observed bidirectional mediation effects (21.1% of H. MicrofluidusThe influence B. Wexlerae was mediated by racetracks).
Coanalysis of lifestyle and metabolome data revealed that around 65.9% of identified metabolite biomarkers are associated with reversible lifestyle changes, highlighting the potential for monitoring the effects of exercise or diet interventions to prevent or treat diabetic results. A high contribution of coffee, common in the Swedish cohort, has reduced the variability of metabolites linked to food, highlighting microbiome adaptations specific to the population. In particular, the propionate of metabolite imidazole was high in the IGT but not validated in the SCAPIS cohort, suggesting a specific variability in the population.
 The thermal card showing the metabolites that overlap involved in the metabolism of amino, lipid and xenobiotic acids (n = 123) in two clinical trials of one or the other of the food interventions (14 days) or the exercise for 1-h (before, 120 and 180 min after the exercise) with these 502 modified metabolites in the prediac and T2D. The inverted responses (y, yes; n, no) by the diet (d) or the exercise (e) or both (b) have been grouped and are represented in distinct colors next to the rows clustering branches. Representative metabolites, including 14 overlapping with fig. 4F, are labeled in red and five others in black. The Wilcoxon row sum test and the analysis of the unidirectional repeated measurement variance were used to identify modified metabolites in cohorts and two longitudinal data sets (padj<0.1), respectively.
The thermal card showing the metabolites that overlap involved in the metabolism of amino, lipid and xenobiotic acids (n = 123) in two clinical trials of one or the other of the food interventions (14 days) or the exercise for 1-h (before, 120 and 180 min after the exercise) with these 502 modified metabolites in the prediac and T2D. The inverted responses (y, yes; n, no) by the diet (d) or the exercise (e) or both (b) have been grouped and are represented in distinct colors next to the rows clustering branches. Representative metabolites, including 14 overlapping with fig. 4F, are labeled in red and five others in black. The Wilcoxon row sum test and the analysis of the unidirectional repeated measurement variance were used to identify modified metabolites in cohorts and two longitudinal data sets (padj<0.1), respectively.
Future conclusions and orientations
The present study reveals the role of the dynamics of microbiome-metabolome in the modification of the homeostasis of human glucose, triggering prediabetic and T2D. It highlights the importance and potential of lifestyle changes, in particular food and exercise, to adjust and monitor these dynamics to obtain optimal health results. The results were validated in GF / Conv-R mice and external cohorts (Israeli, Twinsuk), strengthening their robustness. Optimal advantages probably require the diet and exercise interventions, as modulated by the metabolite specific to lifestyle (for example, branched chain fatty acids have improved with exercise, 7 hoca with a diet).
“Understand the links between the diet, Intestinal microbiotaAnd clinical factors provide valuable information on T2D and highlights the need for various intervention strategies. This resource can provide an increased understanding of how the intestinal microbiota can affect T2D and help identify new targets for diabetes management. »»
Study authors have developed an open access web server (https://omicsdata.org/apps/igt_metabolome/), which will provide future researchers with an easy-to-use platform for exploring the metabolome, meta-analysis and data visualization.